tidymodelsによるtidyな機械学習(その2:Cross Varidation)
はじめに
本記事ではtidymodelsを用いたCross Validationとハイパーパラメータのチューニングについて紹介したいと思います。 なお、tidymodelsの基本的な操作方法については以下の記事をご覧下さい。
前処理
まずは前回の記事と同様、訓練/テストデータの分割と前処理を行います。なお、例によってデータはdiamondsを用います。
# パッケージ library(tidyverse) library(tidymodels) set.seed(42) # 分割 df_split = initial_split(diamonds, p = 0.8) df_train = training(df_split) df_test = testing(df_split) # 前処理レシピ rec = recipe(price ~ ., data = df_train) %>% step_log(price) %>% step_ordinalscore(all_nominal())
Cross Validation
さて、ここからが新しい内容になります。
df_cv = vfold_cv(df_train, v = 10) > df_cv # 10-fold cross-validation # A tibble: 10 x 2 splits id <list> <chr> 1 <split [38.8K/4.3K]> Fold01 2 <split [38.8K/4.3K]> Fold02 3 <split [38.8K/4.3K]> Fold03 4 <split [38.8K/4.3K]> Fold04 5 <split [38.8K/4.3K]> Fold05 6 <split [38.8K/4.3K]> Fold06 7 <split [38.8K/4.3K]> Fold07 8 <split [38.8K/4.3K]> Fold08 9 <split [38.8K/4.3K]> Fold09 10 <split [38.8K/4.3K]> Fold10
tidymodels配下のrsampleパケージにはCross Validationを行うための関数vfold_cv()があるので、これを使います。
> df_cv$splits[[1]] <38837/4316/43153>
splitsを確認します。全43153サンプルを38837の訓練データと4316の検証データに分割していることを示しています。
> df_cv$splits[[1]] %>% analysis() # A tibble: 38,837 x 10 carat cut color clarity depth table price x y z <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl> 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31 3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31 4 0.290 Premium I VS2 62.4 58 334 4.2 4.23 2.63 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75 6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48 7 0.26 Very Good H SI1 61.9 55 337 4.07 4.11 2.53 8 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49 9 0.23 Very Good H VS1 59.4 61 338 4 4.05 2.39 10 0.3 Good J SI1 64 55 339 4.25 4.28 2.73 > df_cv$splits[[1]] %>% assessment() # A tibble: 4,316 x 10 carat cut color clarity depth table price x y z <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl> 1 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47 2 0.3 Very Good J VS2 62.2 57 357 4.28 4.3 2.67 3 0.23 Very Good F VS1 59.8 57 402 4.04 4.06 2.42 4 0.31 Good H SI1 64 54 402 4.29 4.31 2.75 5 0.32 Good H SI2 63.1 56 403 4.34 4.37 2.75 6 0.22 Premium E VS2 61.6 58 404 3.93 3.89 2.41 7 0.3 Very Good I SI1 63 57 405 4.28 4.32 2.71 8 0.24 Premium E VVS1 60.7 58 553 4.01 4.03 2.44 9 0.26 Very Good D VVS2 62.4 54 554 4.08 4.13 2.56 10 0.86 Fair E SI2 55.1 69 2757 6.45 6.33 3.52 # ... with 4,306 more rows
訓練データにはanalysis()で、検証データにはassesment()でアクセスできます。
CVデータが準備できたので、学習と予測を行いましょう。まずは適当なRandom Forestモデルを用意します。
rf = rand_forest(mode = "regression", trees = 50, min_n = 10, mtry = 3) %>% set_engine("ranger", num.threads = parallel::detectCores(), seed = 42)
CVデータに前処理レシピを適用するにはmap()とprepper()を使います。
> df_cv %>% + mutate(recipes = map(splits, prepper, recipe = rec)) # 10-fold cross-validation # A tibble: 10 x 3 splits id recipes * <list> <chr> <list> 1 <split [38.8K/4.3K]> Fold01 <S3: recipe> 2 <split [38.8K/4.3K]> Fold02 <S3: recipe> 3 <split [38.8K/4.3K]> Fold03 <S3: recipe> 4 <split [38.8K/4.3K]> Fold04 <S3: recipe> 5 <split [38.8K/4.3K]> Fold05 <S3: recipe> 6 <split [38.8K/4.3K]> Fold06 <S3: recipe> 7 <split [38.8K/4.3K]> Fold07 <S3: recipe> 8 <split [38.8K/4.3K]> Fold08 <S3: recipe> 9 <split [38.8K/4.3K]> Fold09 <S3: recipe> 10 <split [38.8K/4.3K]> Fold10 <S3: recipe>
prepper()はprep()前のレシピを渡すことで、CV訓練データを用いてprep()したレシピを返してくれます。
次に前処理済みのCVデータでモデルを学習します。
> df_cv %>% + mutate(recipes = map(splits, prepper, recipe = rec), + fitted = map(recipes, ~ fit(rf, price ~ ., data = juice(.)))) # 10-fold cross-validation # A tibble: 10 x 4 splits id recipes fitted * <list> <chr> <list> <list> 1 <split [38.8K/4.3K]> Fold01 <S3: recipe> <fit[+]> 2 <split [38.8K/4.3K]> Fold02 <S3: recipe> <fit[+]> 3 <split [38.8K/4.3K]> Fold03 <S3: recipe> <fit[+]> 4 <split [38.8K/4.3K]> Fold04 <S3: recipe> <fit[+]> 5 <split [38.8K/4.3K]> Fold05 <S3: recipe> <fit[+]> 6 <split [38.8K/4.3K]> Fold06 <S3: recipe> <fit[+]> 7 <split [38.8K/4.3K]> Fold07 <S3: recipe> <fit[+]> 8 <split [38.8K/4.3K]> Fold08 <S3: recipe> <fit[+]> 9 <split [38.8K/4.3K]> Fold09 <S3: recipe> <fit[+]> 10 <split [38.8K/4.3K]> Fold10 <S3: recipe> <fit[+]>
学習モデルを用いて検証用データに対して予測を行います。
# ラッパーを作っておく pred_wrapper = function(split_obj, rec_obj, model_obj, ...) { df_pred = bake(rec_obj, assessment(split_obj)) %>% bind_cols(predict(model_obj, .)) %>% select(price, .pred) return(df_pred) } > df_cv %>% + mutate(recipes = map(splits, prepper, recipe = rec), + fitted = map(recipes, ~ fit(rf, price ~ ., data = juice(.))), + predicted = pmap(list(splits, recipes, fitted), pred_wrapper)) # 10-fold cross-validation # A tibble: 10 x 5 splits id recipes fitted predicted * <list> <chr> <list> <list> <list> 1 <split [38.8K/4.3K]> Fold01 <S3: recipe> <fit[+]> <tibble [4,316 × 2]> 2 <split [38.8K/4.3K]> Fold02 <S3: recipe> <fit[+]> <tibble [4,316 × 2]> 3 <split [38.8K/4.3K]> Fold03 <S3: recipe> <fit[+]> <tibble [4,316 × 2]> 4 <split [38.8K/4.3K]> Fold04 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> 5 <split [38.8K/4.3K]> Fold05 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> 6 <split [38.8K/4.3K]> Fold06 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> 7 <split [38.8K/4.3K]> Fold07 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> 8 <split [38.8K/4.3K]> Fold08 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> 9 <split [38.8K/4.3K]> Fold09 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> 10 <split [38.8K/4.3K]> Fold10 <S3: recipe> <fit[+]> <tibble [4,315 × 2]>
pmap()は入力列が複数の場合のmap()の拡張です。
予測結果を用いて検証用データで精度を計算して完了です。
> df_cv %>% + mutate(recipes = map(splits, prepper, recipe = rec), + fitted = map(recipes, ~ fit(rf, price ~ ., data = juice(.))), + predicted = pmap(list(splits, recipes, fitted), pred_wrapper), + evaluated = map(predicted, metrics, price, .pred)) # 10-fold cross-validation # A tibble: 10 x 6 splits id recipes fitted predicted evaluated * <list> <chr> <list> <list> <list> <list> 1 <split [38.8K/4.3K]> Fold01 <S3: recipe> <fit[+]> <tibble [4,316 × 2]> <tibble [3 × 3]> 2 <split [38.8K/4.3K]> Fold02 <S3: recipe> <fit[+]> <tibble [4,316 × 2]> <tibble [3 × 3]> 3 <split [38.8K/4.3K]> Fold03 <S3: recipe> <fit[+]> <tibble [4,316 × 2]> <tibble [3 × 3]> 4 <split [38.8K/4.3K]> Fold04 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> <tibble [3 × 3]> 5 <split [38.8K/4.3K]> Fold05 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> <tibble [3 × 3]> 6 <split [38.8K/4.3K]> Fold06 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> <tibble [3 × 3]> 7 <split [38.8K/4.3K]> Fold07 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> <tibble [3 × 3]> 8 <split [38.8K/4.3K]> Fold08 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> <tibble [3 × 3]> 9 <split [38.8K/4.3K]> Fold09 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> <tibble [3 × 3]> 10 <split [38.8K/4.3K]> Fold10 <S3: recipe> <fit[+]> <tibble [4,315 × 2]> <tibble [3 × 3]>
ハイパーパラメータのサーチ
Cross Validationで精度を計算する方法がわかったので、これを用いてハイパーパラメータのサーチを行います。
今回はmin_nのみグリッドサーチすることにします。
grid_min_n = c(5, 10, 15) df_result_cv = tibble() # 結果をいれる for (n in grid_min_n) { rf = rand_forest(mode = "regression", trees = 50, min_n = n, mtry = 3) %>% set_engine("ranger", num.threads = parallel::detectCores(), seed = 42) tmp_result = df_cv %>% mutate(recipes = map(splits, prepper, recipe = rec), fitted = map(recipes, ~ fit(rf, price ~ ., data = juice(.))), predicted = pmap(list(splits, recipes, fitted), pred_wrapper), evaluated = map(predicted, metrics, price, .pred)) df_result_cv = df_result_cv %>% bind_rows(tmp_result %>% select(id, evaluated) %>% mutate(min_n = n)) } > df_result_cv %>% + unnest() %>% + group_by(min_n, .metric) %>% + summarise(mean(.estimate)) # A tibble: 9 x 3 # Groups: min_n [?] min_n .metric `mean(.estimate)` <dbl> <chr> <dbl> 1 5 mae 0.0671 2 5 rmse 0.0932 3 5 rsq 0.992 4 10 mae 0.0679 5 10 rmse 0.0940 6 10 rsq 0.991 7 15 mae 0.0686 8 15 rmse 0.0947 9 15 rsq 0.991
どうやらmin_n = 5の精度が最も良さそうです。
このパラメータを用いて訓練データ全体でモデルを再学習し、予測精度を確認します。
parsnipで作ったモデルはupdate()でパラメータを更新できます。
rf_best = update(rf, min_n = 5) rec_preped = rec %>% prep(df_train) fitted = rf_best %>% fit(price ~ ., data = juice(rec_preped)) df_test_baked = bake(rec_preped, df_test) > df_test_baked %>% + bind_cols(predict(fitted, df_test_baked)) %>% + metrics(price, .pred) # A tibble: 3 x 3 .metric .estimator .estimate <chr> <chr> <dbl> 1 rmse standard 0.0905 2 rsq standard 0.992 3 mae standard 0.0665
予測結果と実際の値もプロットして確認します。
df_test_baked %>% bind_cols(predict(fitted, df_test_baked)) %>% ggplot(aes(.pred, price)) + geom_hex() + geom_abline(slope = 1, intercept = 0) + coord_fixed() + scale_x_continuous(breaks = seq(6, 10, 1), limits = c(5.8, 10.2)) + scale_y_continuous(breaks = seq(6, 10, 1), limits = c(5.8, 10.2)) + scale_fill_viridis_c() + labs(x = "Prediction", y = "Truth") + theme_minimal(base_size = 12) + theme(panel.grid.minor = element_blank(), axis.text = element_text(color = "black", size =12))
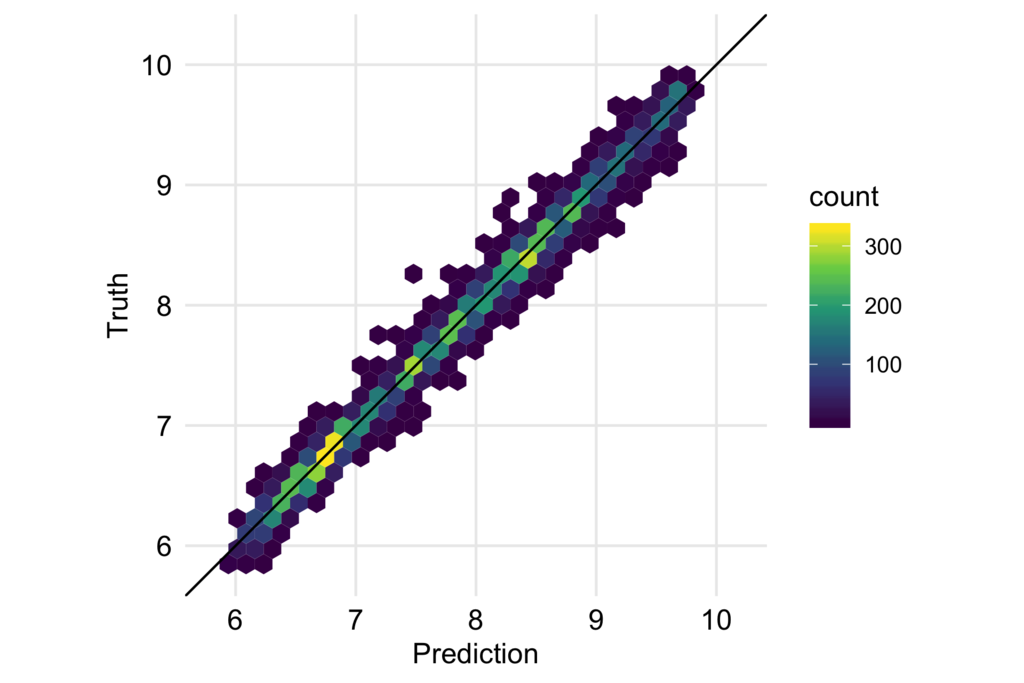
大きく外したり系統的なバイアスも見られないので、うまく予測できていると言えるのではないかと思います。
まとめ
tidymodels配下のパッケージ、特にpurrrとrsampleを用いることでtidyにCross Validationを行うことができました。